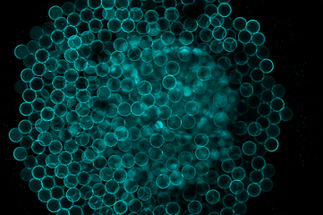
Biomicrofluidic Systems
Bottom-up synthetic biology
The Robinson lab is embedded within the joint project MaxSynBio, sponsored by the Max Planck Society and the German Ministry for Education and Research (BMBF). The overall objective of MaxSynBio is the creation of a minimal cell from functional modules by means of a bottom-up approach to synthetic biology. The functional modules themselves being made from non-living components. The interdisciplinary joint research project involves 9 Max Planck Institutes in the fields of biology, chemistry, physics and engineering sciences. Our group uses lipid vesicles, namely GUVs, as a means to construct artificial cell-like systems. In the same way the cell plasma membrane acts as a barrier to the outside environment, we use GUVs as compartments to encapsulate various biomolecules to enable de novo mimicry of biological processes.
Methodologies and research interests:
- Microfluidic systems: droplets, double emulsion, and vesicle traps
- GUVs: phase-transfer, electroformation, gentle hydration, asymmetric membranes, microfluidic GUVs (mGUVs)
- Single-cell analysis using microfluidics
- LUVs: extrusion
- Phase-separated lipid membranes (domains, lipid raft models)
- Advanced microscopy including confocal, FLIM, FRET, FLIM-FRET, high-speed camera, STED
- Membrane-membrane adhesion (DNA, proteins)
- Membrane proteins including pore proteins and ion channels/transporters
- Membrane fusion
Selected publications:
1.
Yandrapalli, N.; Petit, J.; Bäumchen, O.; Robinson, T.: Surfactant-free production of biomimetic giant unilamellar vesicles using PDMS-based microfluidics. Communications Chemistry 4, 100 (2021)
2.
Shetty, S.; Yandrapalli, N.; Pinkwart, K.; Krafft, D.; Vidaković-Koch, T.; Ivanov, I.; Robinson, T.: Directed signaling cascades in monodisperse artificial eukaryotic cells. ACS Nano 15 (10), S. 15656 - 15666 (2021)
3.
Love, C.; Steinkühler, J.; Gonzales, D.; Yandrapalli, N.; Robinson, T.; Dimova, R.; Tang, D.: Reversible pH responsive coacervate formation in lipid vesicles activates dormant enzymatic reactions. Angewandte Chemie, International Edition in English (2020)
4.
Robinson, T.; Dittrich, P. S.: Observations of membrane domain reorganization in mechanically compressed artificial cells. Chembiochem 20 (20), S. 2666 - 2673 (2019)
5.
Yandrapalli, N.; Robinson, T.: Ultra-high capacity microfluidic trapping of giant vesicles for high-throughput membrane studies. Lab on a Chip 19 (4), S. 626 - 633 (2019)
6.
Schwille, P.; Spatz, J.; Landfester, K.; Bodenschatz, E.; Herminghaus, S.; Sourjik, V.; Erb, T.; Bastiaens, P.; Lipowsky, R.; Hyman, A. et al.; Dabrock, P.; Baret, J.-C.; Vidakovic-Koch, T.; Bieling, P.; Dimova, R.; Mutschler, H.; Robinson, T.; Tang, D.; Wegner, S.; Sundmacher, K.: MaxSynBio ‐ avenues towards creating cells from the bottom up. Angewandte Chemie International Edition
57 (41), S. 13382 - 13392 (2018)
7.
Kubsch, B.; Robinson, T.; Lipowsky, R.; Dimova, R.: Solution asymmetry and salt expand fluid-fluid coexistence regions of charged membranes. Biophysical Journal 110 (12), S. 2581 - 2584 (2016)
8.
Sturzenegger, F.; Robinson, T.; Hess, D.; Dittrich, P. S.: Membranes under shear stress: visualization of non-equilibrium domain patterns and domain fusion in a microfluidic device. Soft Matter 12 (23), S. 5072 - 5076 (2016)
9.
Robinson, T.; Verboket, P. E.; Eyer, K.; Dittrich, P. S.: Controllable electrofusion of lipid vesicles: initiation and analysis of reactions within biomimetic containers. Lab on a Chip 14, S. 2852 - 2859 (2014)
10.
Robinson, T.; Valluri, P.; Kennedy, G.; Sardini, A.; Dunsby, C.; Neil, M. A. A.; Baldwin, G. S.; French, P. M. W.; de Mello, A. J.: Analysis of DNA Binding and Nucleotide Flipping Kinetics Using Two-Color Two-Photon Fluorescence Lifetime Imaging Microscopy. Analytical Chemistry 86 (21), S. 10732 - 10740 (2014)
Multi-compartment lipid membrane systems
Eukaryotic cells have evolved membrane-bound sub-cellular organelles. The advantage of which is to provide spatial organisation of biomolecules, therefore allowing more diverse cellular functionalities.
[mehr]