Unlocking the different tempi of protein synthesis
Scientists of the Max Planck Institutes of Colloids and Interfaces in Potsdam and for Biophysical Chemistry in Göttingen have developed a new method to determine the different codon-specific speeds of protein synthesis in living cells from in-vitro experiments.
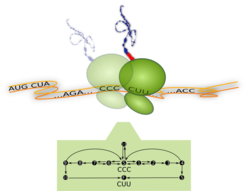
Ribosome translating one codon and corresponding process diagram: Top –The Ribosome (green) is a molecular machine with a diameter of about 23 nm. It moves along the messenger-RNA (orange) and translates it codon by codon (... CCC, CUU,...) into a sequence of amino acids. For each codon, a new amino acid (red) is attached to the growing peptide chain (blue). Bottom – Process diagram with internal reaction pathways (black arrows) of the ribosome translating the codon CCC. The ribosome starts in its internal state S (start), from where it can follow three different reaction pathways. These pathways correspond to the binding of different transfer-RNA molecules (not shown). Translation of the codon is finished as soon as the ribosome reaches the molecular state F (finish) which means that it has moved to the next codon (here CUU). Whenever multiple pathways originate from the same internal state, the ribosome follows them with different probabilities.
The proverb ‘life is motion’ applies both to the macroscopic and the microscopic world. Indeed, if we looked into any living cell with molecular resolution, we could see that cellular components are constantly created, modified, transported and finally degraded. These dynamic processes are driven by a variety of tiny biomolecular machines. An impressive example for such a process is the synthesis of proteins by ribosomes (Fig. 1). The ribosome has a diameter of 23 nanometers and is therefore a true “nanorobot”.
During protein synthesis the ribosome moves along a messenger-RNA, which consists of a chain of codons. It translates the codon sequence of the messenger-RNA into the amino acid sequence of the associated protein. As soon as the ribosome arrives at a new codon it waits for a cognate transfer-RNA loaded with the correct amino acid. During the translation of a single codon, the ribosome follows a series of internal transformations (process diagram, Fig. 1) to finally elongate the growing peptide chain by one amino acid. Afterwards the ribosome moves on to translate the next codon. This process continues until the whole protein has been completed.
Reliable and precise protein synthesis by ribosomes is crucial for all life processes. Not only our cells but also the cells of all other organisms consist for the most part of proteins. When the synthesis of new proteins is inhibited, a cell can no longer work properly and dies. This is actually a well-known strategy to fight bacterial infections: Antibiotics block individual steps of protein synthesis in bacteria, hence killing the microorganisms.
But ribosomes do not work like macroscopic machines. They randomly follow different reaction pathways (process diagram, Fig. 1), which correspond to the binding of different transfer-RNAs. In fact, ribosomes have to examine many different transfer-RNAs in order to find and recognize a correct amino acid. Furthermore, the duration of an individual step of the reaction pathways is not entirely fixed but can be different every time it is performed.
Although ribosomes follow the single reaction steps more or less randomly, they work very reliably and almost never incorporate a wrong amino acid. However, the different reaction pathways with random step times lead to an important consequence: Different codons are translated with different speeds. Scientists of the Max Planck Institutes of Colloids and Interfaces in Potsdam and for Biophysical Chemistry in Göttingen have now studied how much the speed of translation depends on the individual step times and how much it differs for the 61 sense codons in living cells.
Currently it is not possible to measure the individual step times directly in living cells, as we cannot observe the highly complex interior of a cell with molecular resolution. Therefore, experiments are usually carried out in vitro: Ribosomes are isolated from cells and then studied “in the test tube”. “The in vitro behavior is not necessarily the same as in the cell. One could compare this to a plant, which grows much better in nature than on the window sill at home. Now the question rises what we can actually learn from these in vitro experiments?” says Sophia Rudorf, scientist at the MPI of Colloids and Interfaces.
The researchers wanted to bridge the gap between in vitro experiments and translation in real cells. Therefore, they have now combined experimental in vitro data on protein synthesis, which were collected over many years at the MPI in Göttingen, with a new mathematical algorithm that has been developed at the MPI in Potsdam. The algorithm modifies the step times measured in vitro to predict their values in living cells. It does so by first determining all step times, which are theoretically compatible with known properties of the process in the cell. Then, out of all theoretically possible step times, the algorithm selects those that are closest to the step times measured in vitro. In order to do so, the scientists have introduced a new concept, the kinetic distance. The predicted step times of protein synthesis were successfully validated with existing data on speed and accuracy of translation in bacteria. It turned out that the speed of protein synthesis varies a lot for the 61 codons. The slowest codon is translated more than ten times slower than the fastest codon.
The new method allows elucidating the dynamics of protein synthesis in living cells. “This is not only relevant for the investigation of antibiotics. Many diseases arise from genetic mutations, which result in a strongly hampered protein synthesis. The new findings could help us to understand these diseases and their underlying mechanisms”, explains Rudorf. But the new method could also be applied to many other cellular processes such as catalytic activities of enzymes or protein folding. A better understanding of these processes should be highly valuable in order to develop new treatments for a broad variety of diseases.












